Immunohistochemical staining colors separation¶
In this example we separate the immunohistochemical (IHC) staining from the hematoxylin counterstaining. The separation is achieved with the method described in [1], known as “color deconvolution”.
The IHC staining expression of the FHL2 protein is here revealed with Diaminobenzidine (DAB) which gives a brown color.
| [1] | A. C. Ruifrok and D. A. Johnston, “Quantification of histochemical staining by color deconvolution.,” Analytical and quantitative cytology and histology / the International Academy of Cytology [and] American Society of Cytology, vol. 23, no. 4, pp. 291-9, Aug. 2001. |
import matplotlib.pyplot as plt
from skimage import data
from skimage.color import rgb2hed
ihc_rgb = data.immunohistochemistry()
ihc_hed = rgb2hed(ihc_rgb)
fig, axes = plt.subplots(2, 2, figsize=(7, 6))
ax0, ax1, ax2, ax3 = axes.ravel()
ax0.imshow(ihc_rgb)
ax0.set_title("Original image")
ax1.imshow(ihc_hed[:, :, 0], cmap=plt.cm.gray)
ax1.set_title("Hematoxylin")
ax2.imshow(ihc_hed[:, :, 1], cmap=plt.cm.gray)
ax2.set_title("Eosin")
ax3.imshow(ihc_hed[:, :, 2], cmap=plt.cm.gray)
ax3.set_title("DAB")
for ax in axes.ravel():
ax.axis('off')
fig.subplots_adjust(hspace=0.3)
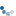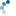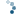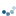

Now we can easily manipulate the hematoxylin and DAB “channels”:
import numpy as np
from skimage.exposure import rescale_intensity
# Rescale hematoxylin and DAB signals and give them a fluorescence look
h = rescale_intensity(ihc_hed[:, :, 0], out_range=(0, 1))
d = rescale_intensity(ihc_hed[:, :, 2], out_range=(0, 1))
zdh = np.dstack((np.zeros_like(h), d, h))
fig, ax = plt.subplots()
ax.imshow(zdh)
ax.set_title("Stain separated image (rescaled)")
ax.axis('off')
plt.show()
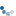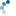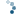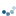

STDOUT
STDERR
Python source code: download (generated using skimage 0.11dev)
IPython Notebook: download (generated using skimage 0.11dev)
aW1wb3J0IG1hdHBsb3RsaWIucHlwbG90IGFzIHBsdAoKZnJvbSBza2ltYWdlIGltcG9ydCBkYXRhCmZyb20gc2tpbWFnZS5jb2xvciBpbXBvcnQgcmdiMmhlZAoKCmloY19yZ2IgPSBkYXRhLmltbXVub2hpc3RvY2hlbWlzdHJ5KCkKaWhjX2hlZCA9IHJnYjJoZWQoaWhjX3JnYikKCmZpZywgYXhlcyA9IHBsdC5zdWJwbG90cygyLCAyLCBmaWdzaXplPSg3LCA2KSkKYXgwLCBheDEsIGF4MiwgYXgzID0gYXhlcy5yYXZlbCgpCgpheDAuaW1zaG93KGloY19yZ2IpCmF4MC5zZXRfdGl0bGUoIk9yaWdpbmFsIGltYWdlIikKCmF4MS5pbXNob3coaWhjX2hlZFs6LCA6LCAwXSwgY21hcD1wbHQuY20uZ3JheSkKYXgxLnNldF90aXRsZSgiSGVtYXRveHlsaW4iKQoKYXgyLmltc2hvdyhpaGNfaGVkWzosIDosIDFdLCBjbWFwPXBsdC5jbS5ncmF5KQpheDIuc2V0X3RpdGxlKCJFb3NpbiIpCgpheDMuaW1zaG93KGloY19oZWRbOiwgOiwgMl0sIGNtYXA9cGx0LmNtLmdyYXkpCmF4My5zZXRfdGl0bGUoIkRBQiIpCgpmb3IgYXggaW4gYXhlcy5yYXZlbCgpOgogICAgYXguYXhpcygnb2ZmJykKCmZpZy5zdWJwbG90c19hZGp1c3QoaHNwYWNlPTAuMyk=
aW1wb3J0IG51bXB5IGFzIG5wCgpmcm9tIHNraW1hZ2UuZXhwb3N1cmUgaW1wb3J0IHJlc2NhbGVfaW50ZW5zaXR5CgojIFJlc2NhbGUgaGVtYXRveHlsaW4gYW5kIERBQiBzaWduYWxzIGFuZCBnaXZlIHRoZW0gYSBmbHVvcmVzY2VuY2UgbG9vawpoID0gcmVzY2FsZV9pbnRlbnNpdHkoaWhjX2hlZFs6LCA6LCAwXSwgb3V0X3JhbmdlPSgwLCAxKSkKZCA9IHJlc2NhbGVfaW50ZW5zaXR5KGloY19oZWRbOiwgOiwgMl0sIG91dF9yYW5nZT0oMCwgMSkpCnpkaCA9IG5wLmRzdGFjaygobnAuemVyb3NfbGlrZShoKSwgZCwgaCkpCgpmaWcsIGF4ID0gcGx0LnN1YnBsb3RzKCkKYXguaW1zaG93KHpkaCkKYXguc2V0X3RpdGxlKCJTdGFpbiBzZXBhcmF0ZWQgaW1hZ2UgKHJlc2NhbGVkKSIpCmF4LmF4aXMoJ29mZicpCnBsdC5zaG93KCk=
