Gabors / Primary Visual Cortex “Simple Cells” from Lena¶
How to build a (bio-plausible) “sparse” dictionary (or ‘codebook’, or ‘filterbank’) for e.g. image classification without any fancy math and with just standard python scientific libraries?
Please find below a short answer ;-)
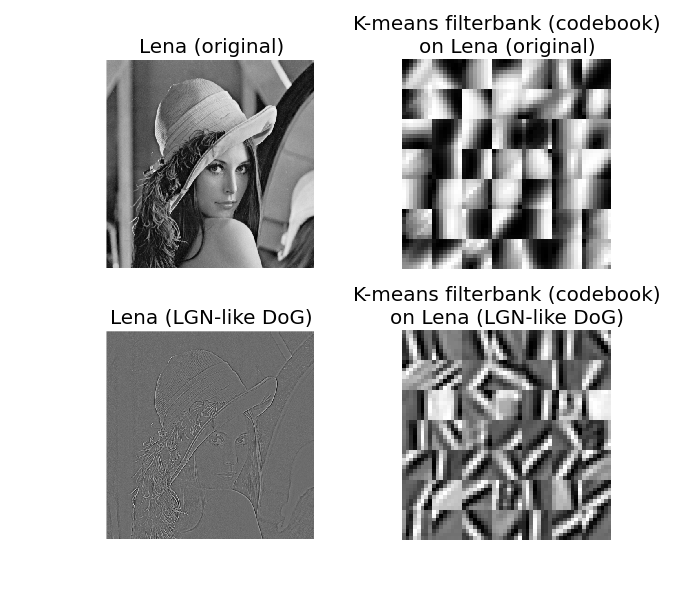
This simple example shows how to get Gabor-like filters [1] using just the famous Lena image. Gabor filters are good approximations of the “Simple Cells” [2] receptive fields [3] found in the mammalian primary visual cortex (V1) (for details, see e.g. the Nobel-prize winning work of Hubel & Wiesel done in the 60s [4] [5]).
Here we use McQueen’s ‘kmeans’ algorithm [6], as a simple biologically plausible hebbian-like learning rule and we apply it (a) to patches of the original Lena image (retinal projection), and (b) to patches of an LGN-like [7] Lena image using a simple difference of gaussians (DoG) approximation.
Enjoy ;-) And keep in mind that getting Gabors on natural image patches is not rocket science.
| [1] | http://en.wikipedia.org/wiki/Gabor_filter |
| [2] | http://en.wikipedia.org/wiki/Simple_cell |
| [3] | http://en.wikipedia.org/wiki/Receptive_field |
| [4] | http://en.wikipedia.org/wiki/K-means_clustering |
| [5] | http://en.wikipedia.org/wiki/Lateral_geniculate_nucleus |
| [6] | D. H. Hubel and T. N., Wiesel Receptive Fields of Single Neurones in the Cat’s Striate Cortex, J. Physiol. pp. 574-591 (148) 1959 |
| [7] | D. H. Hubel and T. N., Wiesel Receptive Fields, Binocular Interaction, and Functional Architecture in the Cat’s Visual Cortex, J. Physiol. 160 pp. 106-154 1962 |
import numpy as np
from scipy.cluster.vq import kmeans2
from scipy import ndimage as ndi
import matplotlib.pyplot as plt
from skimage import data
from skimage import color
from skimage.util.shape import view_as_windows
from skimage.util.montage import montage2d
np.random.seed(42)
patch_shape = 8, 8
n_filters = 49
lena = color.rgb2gray(data.lena())
# -- filterbank1 on original Lena
patches1 = view_as_windows(lena, patch_shape)
patches1 = patches1.reshape(-1, patch_shape[0] * patch_shape[1])[::8]
fb1, _ = kmeans2(patches1, n_filters, minit='points')
fb1 = fb1.reshape((-1,) + patch_shape)
fb1_montage = montage2d(fb1, rescale_intensity=True)
# -- filterbank2 LGN-like Lena
lena_dog = ndi.gaussian_filter(lena, .5) - ndi.gaussian_filter(lena, 1)
patches2 = view_as_windows(lena_dog, patch_shape)
patches2 = patches2.reshape(-1, patch_shape[0] * patch_shape[1])[::8]
fb2, _ = kmeans2(patches2, n_filters, minit='points')
fb2 = fb2.reshape((-1,) + patch_shape)
fb2_montage = montage2d(fb2, rescale_intensity=True)
# --
fig, axes = plt.subplots(2, 2, figsize=(7, 6))
ax0, ax1, ax2, ax3 = axes.ravel()
ax0.imshow(lena, cmap=plt.cm.gray)
ax0.set_title("Lena (original)")
ax1.imshow(fb1_montage, cmap=plt.cm.gray, interpolation='nearest')
ax1.set_title("K-means filterbank (codebook)\non Lena (original)")
ax2.imshow(lena_dog, cmap=plt.cm.gray)
ax2.set_title("Lena (LGN-like DoG)")
ax3.imshow(fb2_montage, cmap=plt.cm.gray, interpolation='nearest')
ax3.set_title("K-means filterbank (codebook)\non Lena (LGN-like DoG)")
for ax in axes.ravel():
ax.axis('off')
fig.subplots_adjust(hspace=0.3)
plt.show()
Python source code: download (generated using skimage 0.10dev)
IPython Notebook: download (generated using skimage 0.10dev)
